Research Interests
Centrosomes are microtubule-organizing centers with diverse functions directing cell division, cell polarization, and cilia formation. The deregulation of centrosomes is associated with developmental disorders, including microcephaly, dwarfism, and ciliopathy. Using multi-disciplinary approaches, we investigate how diverse centrosome function is precisely regulated. We aim to inform mechanisms underlying centrosome regulation and reveal the etiology of centrosome-associated neurodevelopmental diseases.
We are particularly interested in the following directions:
- Recent research uncovers centrosome regulation at the mRNA level. We will investigate how mRNA regulation, mediated by various RNA binding proteins (RBPs), contributes to diverse centrosome functions.
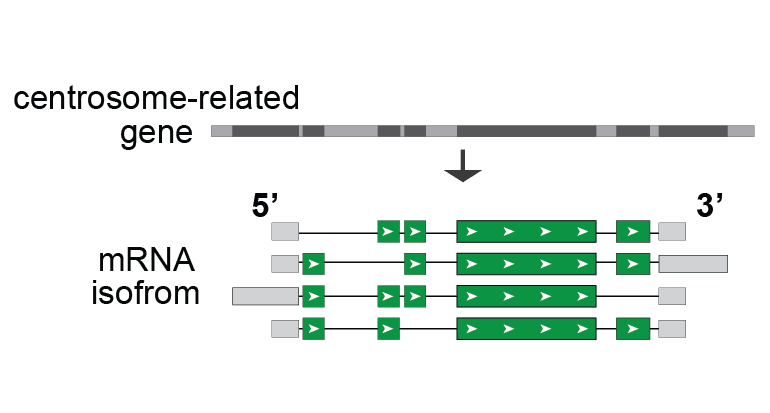
- Most centrosomal genes encode multiple mRNA isoforms, an essential mechanism that diversifies gene products. Using essential centrosome genes as models, we will investigate how isoform expression contributes to diverse centrosome functions.
- Recent studies show that neurodevelopmental disease-relevant RBPs co-exist in the centrosome interactome in human neural stem cells (NSCs) and neurons. We will investigate how centrosome deregulation contributes to neurodevelopmental diseases.

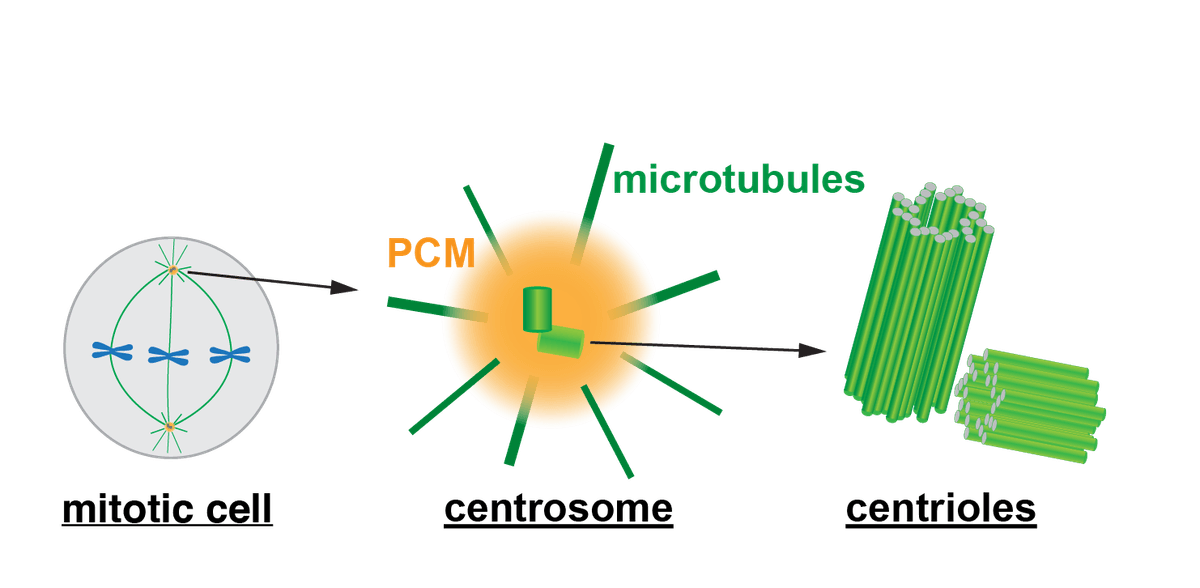
Centrosome structure and function
Functioning as microtubule-organizing centers (MTOC) in most animal cells, centrosomes are composed of a pair of centrioles surrounded by pericentriolar material (PCM). The centrosome is a multi-functional organelle, that directs the formation of mitotic spindles in dividing cells to ensure error-free mitosis, contributes to cell polarity, directs cilia formation, and organizes organelles and traffic intracellular cargo. The deregulation of centrosomes can lead to developmental disorders, including, cancer, microcephaly, and ciliopathy.

Regulation of centrosome-associated mRNA
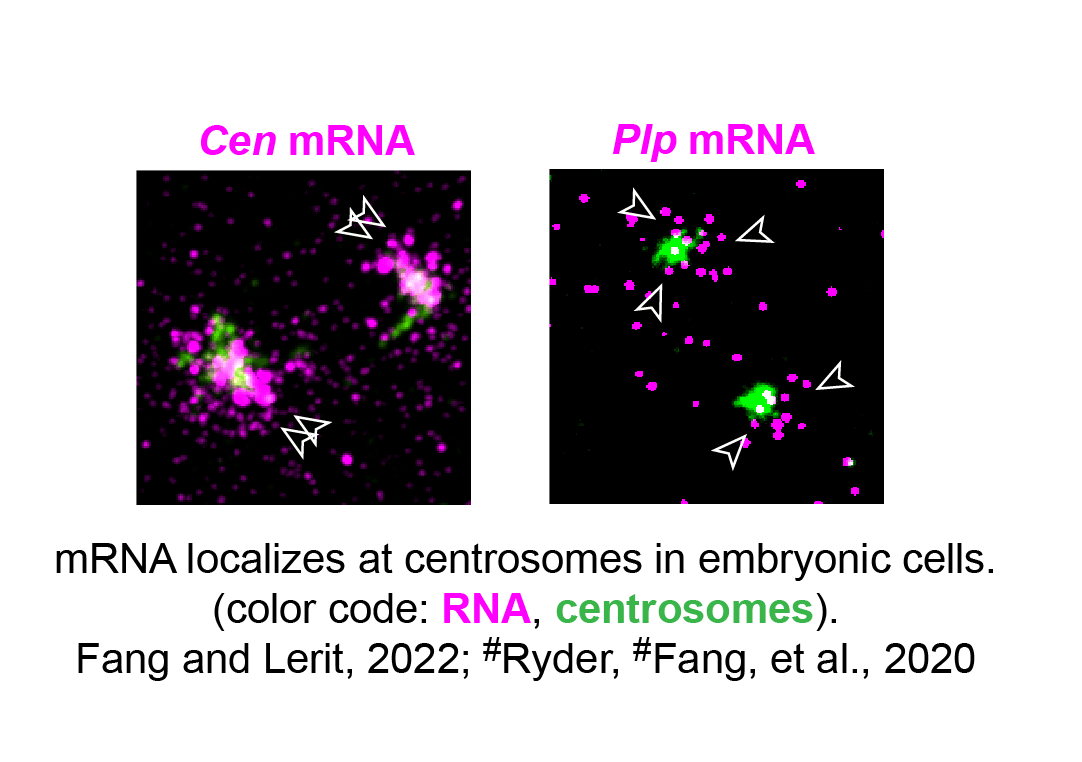
RNA was known to distribute at centrosomes in diverse cell types for decades, indicating that centrosome activity is regulated at the mRNA level. Yet the types of RNA, the underlying localization mechanisms, and their functional roles are largely unknown. Using single-molecule fluorescence in situ hybridization (smFISH), my recent postdoctoral research shows that mRNA encoded by several centrosome-related genes localizes at centrosomes. In addition, using two mRNA: Cen and Plp mRNA as models, we uncover a novel mechanism of centrosome activity controlled by diverse RNA binding proteins, through regulating mRNA localization, mRNA translation, or mRNA polyadenylation (Fang and Lerit, 2022; #Ryder, #Fang, et al., 2020).

Differential mRNA isoform expression & function
Isoform expression is an essential mechanism that diversifies gene products. Most centrosome-related genes encode multiple isoforms. Aberrant isoform expression has gained emerging attention in contributing to human diseases. Yet, how isoform expression dictates diverse centrosome structure/function remains a significant knowledge gap. The Fang lab will employ key centrosome-related genes as models to investigate how isoform expression contributes to centrosome heterogeneity across diverse cell types. (This project is currently supported by an NIGMS K99/R00 award).
Lab Members
Welcome to the Fang lab!

Junnan Fang, Ph.D. Principle Investigator
Assistant Professor of Physiology and Cell Biology
email: fang.1142@osu.edu
Dr. Fang received her Ph.D. in Biochemistry and Molecular Biology from the Institute of Biophysics, Chinese Academy of Sciences. After postdoctoral fellow training at Emory University, she joined The Ohio State University as an Assistant Professor in January 2025. Dr. Fang is a recipient of the NIGMS K99/R00 Pathway to Independence Award. In her leisure time, she enjoys reading, hiking, and traveling.

Danni Li
Research Assistant
li.16303@osu.edu
Danni received her Master degree in Medicine from Guangxi Medical University.

Sarah Liu
Undergraduate student
liu.10009@buckeyemail.osu.edu
Sarah is a fourth-year undergraduate student at Ohio State studying Health Sciences and is completing a minor in Pharmaceutical Sciences. She plans to graduate in the Spring of 2026 and begin applying to medical school. Outside of the lab, she enjoys playing guitar, reading, and photography.

Lauren Jablonski
Undergraduate student
jablonski.66@buckeyemail.osu.edu
Lauren is a undergraduate student at Ohio State on the pre-med track.

Nathan Eads
Undergraduate student
eads.49@buckeyemail.osu.edu
Nathan is a 3rd year undergraduate student majoring in Biochemistry at OSU.
Lab Alumni
Ellie Manning, B.S. (CREATES Undergraduate Program, 2025) | Current: Kenyon College senior student
Ye Zhou, Ph.D. (Postdoctoral Fellow, 2025)
Lab News
2025.10
Danni, Ye, and Sarah attend their first Rustbelt RNA conference and present posters.
MCDB graduate student, Matt Janik, started a rotation in the lab. Welcome!
2025. 09
Nathan joined the lab for reseearch training.
OSBP Graduate student, Ivy Mordick, started a rotation in the lab. Welcome!
2025.07
Dr. Ye Zhou, our new postdoc safely arrived and joined the lab, welcome!
2025.07
We received the NoA of R00 grant!
2025.06
Lauren joined the lab for summer research training, welcome!
2025.05
OSU CREATES Undergraduate Program student, Ellie Manning, joined the lab for a 10-week summer research training, welcome!
2025.05
Sarah joined the lab for summer research training, welcome!
2025.05
Danni joined the lab as research assistant, welcome!
2025.04
OSBP Graduate Student, Gabby Javellana, started a rotation in the lab, welcome!
2025.04
The lab passed EHS inspection and is ready to run.
2025.02
Fly stations were set up.
2025.01
The Fang Lab was officially opened.
Lab Moments




Publications
A full list of publications can be found at Bibliography
- Fang, J.#, Tian, R.#, Quintanilla, M.,Beach, J., Lerit, D.A. The PCM scaffold enables RNA localization to centrosomes. Mol Biol Cell. 2025 June; 36 (6). PMID:12206496.
- #Equal contributors.
- Brockett, J.S., Manalo, T., Zein-Sabatto, H.,Lee, J., Fang, J., Chu, P., Feng, H., Patil, D., Davidson, P., Ogan, K., Master, V.A.,Pattaras, J.G., Roberts, D.L., Bergquist, S.H., Reyna, M. A., Petros, J.A., Lerit, D.A., Arnold, R.S. A missense SNP in the tumor suppressor SETD2 reduces H3K36me3 and mitotic spindle integrity in Drosophila. Genetics. 2024 Apr; 226 (4). PMID:38290049.
- Fang, J., Lerit, D.A. Orb-dependent polyadenylation contributes to PLP expression and centrosome scaffold assembly. Development. 2022 Jul 1;149(13). PMID: 35661190.
- Featured Cover Article. July 2022 Issue.
- #Ryder, P., #Fang, J.,Lerit, D.A. centrocortin RNA localization to centrosomes is regulated by FMRP and facilitateserror-free mitosis. J. Cell. Biol. 2020 Dec 7; 219(12). PMID: 33196763.
- #Equal contributors.
- Fang, J., Lerit, D.A. (2020) DrosophilaPericentrin-like protein promotes the formation of primordial germ cells. Genesis. 2020 Mar; 58(3-4):e23347. PMID:31774613.
- Featured Cover Article. March-April 2020 Issue.
- Fang, J., Jing, X., Lu, G., Jing, X., Xu, Y., Gong P. (2019) Crystallographic snapshots of the Zika virus NS3helicase helps visualize the reactant water replenishment. ACS Infect Dis. Feb 8;5(2):177-183. PMID: 30672289.
- Benner, L., Castro, E.A., Whitworth, C.,Venken, K.J.T., Yang, H., Fang, J., Oliver, B., Cook, K.R., and Lerit,D.A. (2019) Drosophila Heterochromatin stabilization requires the zinc-finger proteinSmall Ovary. Genetics. 2019 Nov;213(3):877-895. PMID: 31558581.
- Fang, Q., Chen, P., Wang, M., Fang, J., Yang, N., Li, G., Xu,RM. (2016) Human cytomegalovirus IE1protein alters the higher-order chromatin structure by targeting the acidic patch of thenucleosome. eLife. Jan 26e11911. PMID: 26812545.
- Fang, J., Liu, Y., Wei, Y., Deng, W., Yu, Z., Huang, L., Teng, Y., Yao, T., You, Q., Ruan, H., Chen, P., Xu, RM.,Li, G. (2015) Structural Transitions of Centromeric Chromatin Coordinate the CellCycle-dependent Recruitment of CENP-N. Genes& Development. 29 (10):1058-73. PMID: 25943375.
- Yu, Z., Zhou, X., Wang, W., Deng, W., Fang, J., Hu, H., Wang Z., Li,S., Cui, L., Shen, J., Zhai, L., Peng, S.,Wong, J., Dong, S., Yuan, Z., Ou, G., Zhang, X., Xu, P., Lou, J., Yang, N., Chen, P., Xu, RM.,Li, G. (2015) Dynamic phosphorylation of Ser68 orchestrates the cellcycle-dependent deposition of CENP-A at centromeres. Developmental Cell. 32: 68-81. PMID:25556658.
We are hiring at all levels!
NIH-funded Postdoctoral fellow positions are available.
Interested applicants are encouraged to directly email Dr. Fang (fang.1142@osu.edu) with the following documents: a brief statement of research interest, an updated CV, and contact details of three references.
PhD applicants: Please apply through the PhD programs at OSU including, MCDB program link here, OSBP program link here. BSGP program link here. You are also welcome to directly email Dr. Fang to discuss research interests.
Training opportunities
In the Fang lab, you'll have the exciting opportunities to publish in prestigious journals, draft and submit your grants, present your research at international conferences, and receive formal & tailored career development training.
Collaborations/Teamwork
We place a strong emphasis on collaboration and teamwork.
Leadership roles, expectations, and contributions will be discussed with all team members at the start of each project, with periodic reviews as the project progresses. Effective communication is essential to our success.
Work/Life Balance
Science is both rewarding and challenging. Maintaining a healthy work-life balance is essential for preserving enthusiasm and productivity. We encourage a balanced approach that allows for dedicated time to both scientific work and personal interests.
Commitment to Inclusion
The Fang lab welcomes students/scholars interested in bioinformatics, genetics, cell biology, RNA biology, and centrosomes regardless of their background.
We are committed to fostering a supportive and inclusive environment within the laboratory and across the broader academic community.
Contact
You are welcome to reach out. You + us = awesome.